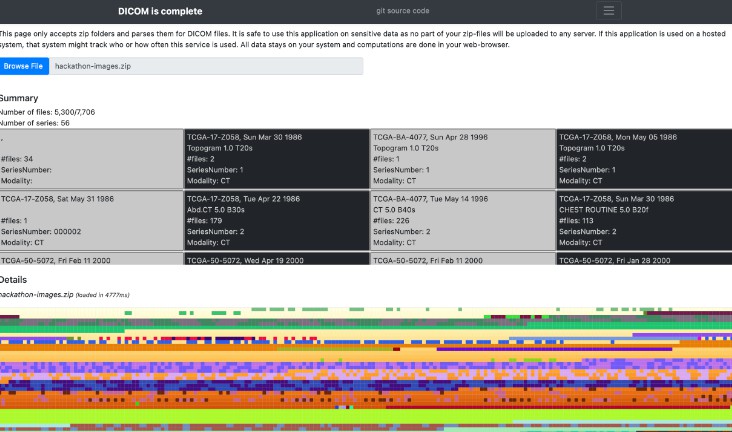
DICOM is complete
A DICOM meta-data viewer that is safe to use in a hospital – based on Javascript, the DICOM-Meta-Data-Viewer.
Create a DICOM file from thin-air (and DCMTK)
mkdir DICOM; cd DICOM
touch empty.dump
dump2dcm empty.dump one.dcm
Add more tags to the DICOM file:
dcmdump one.dcm > step2.dump
echo "(0010,0010) PN [WORKSHOP01]" >> step2.dump
echo "(0010,0020) LN [WORKSHOP01]" >> step2.dump
echo "(0020,000D) UI [1.3.6.1.4.1.45037.5.2.1.987655444]" >> step2.dump
echo "(0020,000E) UI [1.3.6.1.4.1.45037.5.2.1.123456789]" >> step2.dump
dump2dcm step2.dump two.dcm
Add an image informationn:
echo "P1\n10 10\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n0 0 0 0 0 0 0 0 0 0\n" > image.pnm
convert image.pnm image.jpg
img2dcm --series-from two.dcm image.jpg three.dcm
Here is the meta-data from the resulting DICOM image:
> dcmdump three.dcm
# Dicom-File-Format
# Dicom-Meta-Information-Header
# Used TransferSyntax: Little Endian Explicit
(0002,0000) UL 196 # 4, 1 FileMetaInformationGroupLength
(0002,0001) OB 00\01 # 2, 1 FileMetaInformationVersion
(0002,0002) UI =SecondaryCaptureImageStorage # 26, 1 MediaStorageSOPClassUID
(0002,0003) UI [1.2.276.0.7230010.3.1.4.0.15096.1663134574.193501] # 50, 1 MediaStorageSOPInstanceUID
(0002,0010) UI =JPEGFullProgression:Non-hierarchical:Process10+12 # 22, 1 TransferSyntaxUID
(0002,0012) UI [1.2.276.0.7230010.3.0.3.6.6] # 28, 1 ImplementationClassUID
(0002,0013) SH [OFFIS_DCMTK_366] # 16, 1 ImplementationVersionName
# Dicom-Data-Set
# Used TransferSyntax: JPEG Full Progression, Non-hierarchical, Process 10+12
(0008,0005) CS [ISO_IR 100] # 10, 1 SpecificCharacterSet
(0008,0016) UI =SecondaryCaptureImageStorage # 26, 1 SOPClassUID
(0008,0018) UI [1.2.276.0.7230010.3.1.4.0.15096.1663134574.193501] # 50, 1 SOPInstanceUID
(0008,0020) DA (no value available) # 0, 0 StudyDate
(0008,0030) TM (no value available) # 0, 0 StudyTime
(0008,0050) SH (no value available) # 0, 0 AccessionNumber
(0008,0064) CS [WSD] # 4, 1 ConversionType
(0008,0070) LO (no value available) # 0, 0 Manufacturer
(0008,0090) PN (no value available) # 0, 0 ReferringPhysicianName
(0010,0010) PN [WORKSHOP01] # 10, 1 PatientName
(0010,0020) LO [WORKSHOP01] # 10, 1 PatientID
(0010,0030) DA (no value available) # 0, 0 PatientBirthDate
(0010,0040) CS (no value available) # 0, 0 PatientSex
(0020,000d) UI [1.3.6.1.4.1.45037.5.2.1.123456789] # 34, 1 StudyInstanceUID
(0020,000e) UI [1.3.6.1.4.1.45037.5.2.1.987655444] # 34, 1 SeriesInstanceUID
(0020,0010) SH (no value available) # 0, 0 StudyID
(0020,0011) IS (no value available) # 0, 0 SeriesNumber
(0020,0013) IS (no value available) # 0, 0 InstanceNumber
(0020,0020) CS (no value available) # 0, 0 PatientOrientation
(0028,0002) US 1 # 2, 1 SamplesPerPixel
(0028,0004) CS [MONOCHROME2] # 12, 1 PhotometricInterpretation
(0028,0010) US 10 # 2, 1 Rows
(0028,0011) US 10 # 2, 1 Columns
(0028,0100) US 8 # 2, 1 BitsAllocated
(0028,0101) US 8 # 2, 1 BitsStored
(0028,0102) US 7 # 2, 1 HighBit
(0028,0103) US 0 # 2, 1 PixelRepresentation
(0028,2110) CS [01] # 2, 1 LossyImageCompression
(0028,2114) CS [ISO_10918_1] # 12, 1 LossyImageCompressionMethod
(7fe0,0010) OB (PixelSequence #=2) # u/l, 1 PixelData
(fffe,e000) pi (no value available) # 0, 1 Item
(fffe,e000) pi ff\d8\ff\db\00\43\00\03\02\02\03\02\02\03\03\03\03\04\03\03\04\05... # 326, 1 Item
(fffe,e0dd) na (SequenceDelimitationItem) # 0, 0 SequenceDelimitationItem
Changing an existing DICOM file is easy as well with dcmodify:
> dcmdump +P StudyDescription PI.1.3.6.1.4.1.14519.5.2.1.6450.9002.927456543152011698231718997971
(0008,1030) LO [PTCTSKTHPET / CT TUMOR] # 22, 1 StudyDescription
> dcmodify -m "StudyDescription=I changed that" PI.1.3.6.1.4.1.14519.5.2.1.6450.9002.927456543152011698231718997971
> dcmdump +P StudyDescription PI.1.3.6.1.4.1.14519.5.2.1.6450.9002.927456543152011698231718997971
(0008,1030) LO [I changed that] # 14, 1 StudyDescription
Send DICOM around
In order to forward DICOM files to a folder you just need a Sender and a Receiver:
Sender
DCMDICTPATH=/usr/local/Cellar/dcmtk/3.6.6_1/share/dcmtk/dicom.dic
storescp -v \
--aetitle HAUKE \
--exec-on-reception "myScript.sh '#a' '#c' '#r' '#p' '#f’" \
--sort-on-study-uid scp \
--output-directory "/tmp/dicom/" \
11112
Whereas the storescp call is sufficient for receiving and sorting DICOM files into different folders the (data-transfer-station)[https://github.com/HaukeBartsch/data-transfer-station] project provides a solution with additional features:
- sorting of DICOM files into Study/Series/ folders
- creation of JSON structure files that summarize meta-data from DICOM
- classification algorithm for received DICOM files into classes such as T1, T2, etc.
Receiver
cd /to/where/the/data/is/you/want/to/send
storescu -v -nh -aet me -aec HAUKE +r +sd localhost 11112 .
Putting it all together
The sender (SCU) will forward us a copy of each DICOM object. The receiver (SCP) will store it on disk and call our script “myScript.sh”. Here an content of an example myScript.sh:
#/usr/bin/env bash
# We are called for each file once
echo "We got this job: $*"
# Idea
# Create a file for each StudyInstanceUID. Later check how old that file is.
# If the file is older than say 16sec - assume all data arrived and we can
# start processing.
StudyPath="$4"
echo "Study path is: ${StudyPath}"
touchFiles="/tmp/arrived"
# create a directory to remember when we last received an image
if [ ! -d ${touchFiles} ]; then
mkdir -p "${touchFiles}"
fi
StudyInstanceUID=`basename "${StudyPath}"`
# create an empty file, filename is "scp_" + StudyInstanceUID
echo "create touch file: ${touchFiles}/${StudyInstanceUID}"
touch "${touchFiles}/${StudyInstanceUID}"
As a result a file will be created in the /tmp/arrived folder on the receiver’s machine. We can check how new that file is. If its older than 16 seconds we did not receive a new DICOM file for that study in the past 16 seconds. We can assume sending is done and we can react to that by triggering a processing step.
Here an example command line that just lists the study folder names as soon as a study arrives.
watch -n 2 find /tmp/arrived/ -type f -not -newermt \'-16 seconds\'